-
摘要: 地中海贫血又称珠蛋白生成障碍性贫血,是一类具有致死性和致残性的遗传性血液病。地中海贫血目前没有理想的治疗方法,在地中海贫血的防治中,用产前诊断的方式来阻止重型地中海贫血患儿的出生十分重要。地中海贫血目前的基因诊断方式仅能检查出常见的23种突变,而基因测序能对复杂及未知的基因型进行高灵敏度的检测,且已经广泛应用于地中海贫血少见基因型的检测中,在胚胎移植前诊断和无创产前诊断中也有应用。文章基于Sanger、NGS、TGS测序技术的原理、优缺点,对基因测序在地中海贫血诊断中的应用价值做一综述。并基于各代测序的特点,做测序方法选择的建议,作为现行筛查诊断策略的补充。Abstract: Thalassemia, also referred to as globinopathy, is a hereditary blood disorder characterized by its potential lethality and disability. Currently, there is no ideal treatment for thalassemia. In the prevention and treatment of thalassemia, reducing the birth rate of children with severe thalassemia by prenatal diagnosis is particularly important. The existing genetic diagnostic technology for thalassemia can only detect 23 determined common mutations. Gene sequencing can detect complex and unknown genotypes with high sensitivity, and has been widely used in the detection of thalassemia. What's more, it is also used in pre-embryo transfer diagnosis and non-invasive prenatal diagnosis. Based on the principle, advantages and disadvantages of Sanger, NGS and TGS sequencing technology, this paper reviews the application value of gene sequencing in the diagnosis of thalassemia. Above all, the selection of sequencing methods is recommended as a supplement to the current screening and diagnostic strategy.
-
Key words:
- thalassemia /
- sequencing technology /
- prenatal diagnosis /
- screening strategy
-

-
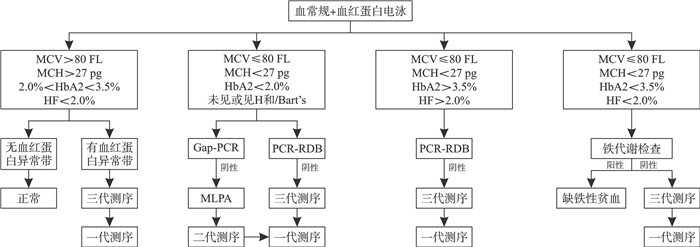
表 1 各代测序在地中海贫血检测中的优缺点
测序技术 一代测序 NGS TGS 读长 700~1 000 bp 100~150 bp 高达几十kb 错误率 0.001% <1% 13%~20% 数据通量 低 较高 很高 高度同源序列的检测 不准确 不准确 准确 结构变异检出能力 较差 较差 较高 是否需要PCR 需要 需要 不需要 在地中海贫血检测中适用场景 金标准,小片段测序 缺失断点检测 结构变异 -
[1] Vijian D, Wan Ab Rahman WS, Ponnuraj KT, et al. Molecular detection of alpha thalassemia: a review of prevalent techniques[J]. Medeni Med J, 2021, 36(3): 257-269.
[2] Wang R, Ma XH, Qin ZZ, et al. Global, regional, and national burden of thalassemia during 1990-2019: a systematic analysis of the Global Burden of Disease Study 2019[J]. Pediatr Blood Cancer, 2024, 71(9): e31177. doi: 10.1002/pbc.31177
[3] 中华医学会围产医学分会, 中华医学会妇产科学分会产科学组. 地中海贫血妊娠期管理专家共识[J]. 中华围产医学杂志, 2020, 23(9): 577-584.
[4] 北京天使妈妈慈善基金会, 北京师范大学中国公益研究院. 中国地中海贫血蓝皮书: 中国地中海贫血防治状况调查报告(2020)[M]. 北京: 中国社会出版社, 2021: 10-15.
[5] 吕建新, 王晓春. 临床分子生物学检验技术[M]. 北京: 人民卫生出版社, 2015: 170-171.
[6] Tantiworawit A, Kamolsripat T, Piriyakhuntorn P, et al. Survival and causes of death in patients with alpha and beta-thalassemia in Northern Thailand[J]. Ann Med, 2024, 56(1): 2338246. doi: 10.1080/07853890.2024.2338246
[7] Kyle Cromer M, Camarena J, Martin RM, et al. Gene replacement of α-globin with β-globin restores hemoglobin balance in β-thalassemia-derived hematopoietic stem and progenitor cells[J]. Nat Med, 2021, 27(4): 677-687. doi: 10.1038/s41591-021-01284-y
[8] Farmakis D, Porter J, Taher A, et al. 2021 thalassaemia international federation guidelines for the management of transfusion-dependent thalassemia[J]. Hemasphere, 2022, 6(8): e732. doi: 10.1097/HS9.0000000000000732
[9] Jiang F, Zhou JY, Zuo LD, et al. Utilization of multiple genetic methods for prenatal diagnosis of rare thalassemia variants[J]. Front Genet, 2023, 14: 1208102. doi: 10.3389/fgene.2023.1208102
[10] Wang G, Huang HY, Chen L, et al. Characterization of a novel 8.2 kb deletion causing beta-thalassemia[J]. Clin Biochem, 2024, 133-134: 110832. doi: 10.1016/j.clinbiochem.2024.110832
[11] Fan DM, Yang X, Huang LM, et al. Simultaneous detection of target CNVs and SNVs of thalassemia by multiplex PCR and next-generation sequencing[J]. Mol Med Rep, 2019, 19(4): 2837-2848.
[12] Bortolin S, Black M, Modi H, et al. Analytical validation of the tag-it high-throughput microsphere-based universal array genotyping platform: application to the multiplex detection of a panel of thrombophilia-associated single-nucleotide polymorphisms[J]. Clin Chem, 2004, 50(11): 2028-2036. doi: 10.1373/clinchem.2004.035071
[13] 秦丹卿, 姚翠泽, 王继成, 等. PCR-流式荧光杂交用于8005例地中海贫血产前基因诊断的回顾性分析[J]. 中华检验医学杂志, 2022, 45(5): 483-487.
[14] Eid OM, Eid MM, Farid M, et al. MLPA as a genetic assay for the prenatal diagnosis of common aneuploidy: the first Egyptian experience[J]. J Genet Eng Biotechnol, 2022, 20(1): 112. doi: 10.1186/s43141-022-00402-8
[15] Yuregir OO, Ayaz A, Yalcintepe S, et al. Detection of α-thalassemia by using multiplex ligation-dependent probe amplification as an additional method for rare mutations in southern Turkey[J]. Indian J Hematol Blood Transfus, 2016, 32(4): 454-459. doi: 10.1007/s12288-015-0617-z
[16] Schouten J, van Vught P, Galjaard RJ. Multiplex ligation-dependent probe amplification(MLPA)for prenatal diagnosis of common aneuploidies[J]. Methods Mol Biol, 2019, 1885: 161-170.
[17] Luo SQ, Chen XY, Zeng DY, et al. The value of single-molecule real-time technology in the diagnosis of rare thalassemia variants and analysis of phenotype-genotype correlation[J]. J Hum Genet, 2022, 67(4): 183-195.
[18] Sanger F, Air GM, Barrell BG, et al. Nucleotide sequence of bacteriophage phi X174 DNA[J]. Nature, 1977, 265(5596): 687-695.
[19] Kircher M, Kelso J. High-throughput DNA sequencing-concepts and limitations[J]. BioEssays, 2010, 32(6): 524-536.
[20] Hassan S, Bahar R, Johan MF, et al. Next-generation sequencing(NGS)and third-generation sequencing(TGS)for the diagnosis of thalassemia[J]. Diagnostics(Basel), 2023, 13(3): 373.
[21] Hogner S, Lundman E, Strand J, et al. Newborn genetic screening-still a role for Sanger sequencing in the era of NGS[J]. Int J Neonatal Screen, 2023, 9(4): 67.
[22] Shooter C, Rooks H, Thein SL, et al. Next generation sequencing identifies a novel rearrangement in the HBB cluster permitting to-the-base characterization[J]. Hum Mutat, 2015, 36(1): 142-150.
[23] Aziz MA, Khan WA, Banu B, et al. Prenatal diagnosis and screening of thalassemia mutations in Bangladesh: presence of rare mutations[J]. Hemoglobin, 2020, 44(6): 397-401.
[24] Krier JB, Kalia SS, Green RC. Genomic sequencing in clinical practice: applications, challenges, and opportunities[J]. Dialogues Clin Neurosci, 2016, 18(3): 299-312.
[25] Lin YM, Zheng QZ, Zheng TW, et al. Expanded newborn screening for inherited metabolic disorders and genetic characteristics in a southern Chinese population[J]. Clin Chim Acta, 2019, 494: 106-111.
[26] Maiga H, Morrison RD, Duffy PE. Sanger sequencing and deconvolution of polyclonal infections: a quantitative approach to monitor drug-resistant Plasmodium falciparum[J]. EBioMedicine, 2024, 103: 105115.
[27] Mandlik JS, Patil AS, Singh S. Next-generation sequencing(NGS): platforms and applications[J]. J Pharm Bioallied Sci, 2024, 16(Suppl 1): S41-S45.
[28] Lee JY. The principles and applications of high-throughput sequencing technologies[J]. Dev Reprod, 2023, 27(1): 9-24.
[29] Zhu N, Zhang DY, Wang WL, et al. A novel coronavirus from patients with pneumonia in China, 2019[J]. N Engl J Med, 2020, 382(8): 727-733.
[30] Midha MK, Wu MC, Chiu KP. Long-read sequencing in deciphering human genetics to a greater depth[J]. Hum Genet, 2019, 138(11-12): 1201-1215.
[31] Xu LP, Mao AP, Liu H, et al. Long-molecule sequencing: a new approach for identification of clinically significant DNA variants in α-thalassemia and β-thalassemia carriers[J]. J Mol Diagn, 2020, 22(8): 1087-1095.
[32] Ling XT, Wang CH, Li LL, et al. Third-generation sequencing for genetic disease[J]. Clin Chim Acta, 2023, 551: 117624.
[33] Ardui S, Ameur A, Vermeesch JR, et al. Single molecule real-time(SMRT)sequencing comes of age: applications and utilities for medical diagnostics[J]. Nucleic Acids Res, 2018, 46(5): 2159-2168.
[34] Rhoads A, Au KF. PacBio sequencing and its applications[J]. Genomics Proteomics Bioinformatics, 2015, 13(5): 278-289.
[35] van Dijk EL, Jaszczyszyn Y, Naquin D, et al. The third revolution in sequencing technology[J]. Trends Genet, 2018, 34(9): 666-681.
[36] Zhuang JL, Chen CN, Fu WY, et al. Third-generation sequencing as a new comprehensive technology for identifying rare α-and β-globin gene variants in thalassemia alleles in the Chinese population[J]. Arch Pathol Lab Med, 2023, 147(2): 208-214.
[37] Long J, Sun L, Gong FF, et al. Third-generation sequencing: a novel tool detects complex variants in the α-thalassemia gene[J]. Gene, 2022, 822: 146332.
[38] Hoff KJ. The effect of sequencing errors on metagenomic gene prediction[J]. BMC Genomics, 2009, 10: 520.
[39] Rieber N, Zapatka M, Lasitschka B, et al. Coverage bias and sensitivity of variant calling for four whole-genome sequencing technologies[J]. PLoS One, 2013, 8(6): e66621.
[40] van Dijk EL, Auger H, Jaszczyszyn Y, et al. Ten years of next-generation sequencing technology[J]. Trends Genet, 2014, 30(9): 418-426.
[41] Watson M, Warr A. Errors in long-read assemblies can critically affect protein prediction[J]. Nat Biotechnol, 2019, 37(2): 124-126.
[42] Ren ZM, Li WJ, Xing ZH, et al. Detecting rare thalassemia in children with Anemia using third-generation sequencing[J]. Hematology, 2023, 28(1): 2241226.
[43] Bashyam MD, Bashyam L, Savithri GR, et al. Molecular genetic analyses of beta-thalassemia in South India reveals rare mutations in the beta-globin gene[J]. J Hum Genet, 2004, 49(8): 408-413.
[44] Gallienne AE, Iberson NM, Dréau HM, et al. Characterization of a novel deletion causing beta-thalassemia major in an Afghan family[J]. Hemoglobin, 2010, 34(1): 110-114.
[45] de Koning TJ, Jongbloed JDH, Sikkema-Raddatz B, et al. Targeted next-generation sequencing panels for monogenetic disorders in clinical diagnostics: the opportunities and challenges[J]. Expert Rev Mol Diagn, 2015, 15(1): 61-70.
[46] Zhuang JL, Zhang N, Zheng Y, et al. Molecular characterization of similar Hb Lepore Boston-Washington in four Chinese families using third generation sequencing[J]. Sci Rep, 2024, 14(1): 9966.
[47] Xu Z, Hu LP, Liu YY, et al. Comparison of third-generation sequencing and routine polymerase chain reaction in genetic analysis of thalassemia[J]. Arch Pathol Lab Med, 2024, 148(3): 336-344.
[48] 阳彦, 刘艳秋, 陆清, 等. 基于高通量测序的单体型分析在地中海贫血-HLA配型的植入前遗传学诊断中的应用[J]. 中华医学遗传学杂志, 2019, 36(11): 1090-1093.
[49] 何天文, 卢建, 陈创奇, 等. 采用二代测序对α/β复合型地中海贫血夫妇进行胚胎植入前遗传学诊断[J]. 中国实验血液学杂志, 2021, 29(4): 1275-1279.
[50] Erlich HA, López-Peña C, Carlberg KT, et al. Noninvasive prenatal test for β-thalassemia and sickle cell disease using probe capture enrichment and next-generation sequencing of DNA in maternal plasma[J]. J Appl Lab Med, 2022, 7(2): 515-531.
[51] Jiang FM, Liu WQ, Zhang LM, et al. Noninvasive prenatal testing for β-thalassemia by targeted nanopore sequencing combined with relative haplotype dosage(RHDO): a feasibility study[J]. Sci Rep, 2021, 11(1): 5714.
[52] 冯宝莹, 韦洁, 黄秀宁, 等. 广西出生缺陷预防控制现状及发展策略[J]. 广西医学, 2024, 46(1): 1-9.
[53] 罗冰星, 翁俊岭, 乔静怡, 等. 国内外地中海贫血防控政策对比及启示[J]. 中国初级卫生保健, 2023, 37(12): 24-27.
[54] Rigter T, Henneman L, Kristoffersson U, et al. Reflecting on earlier experiences with unsolicited findings: points to consider for next-generation sequencing and informed consent in diagnostics[J]. Hum Mutat, 2013, 34(10): 1322-1328.
[55] Bunnik EM, Dondorp WJ, Bredenoord AL, et al. Mainstreaming informed consent for genomic sequencing: a call for action[J]. Eur J Cancer, 2021, 148: 405-410.
-

计量
- 文章访问数: 42
- 施引文献: 0



 下载:
下载: