Feasibility study of Y-chromosome mini-STRs-based next-generation sequencing for non-invasive prenatal paternity testing
-
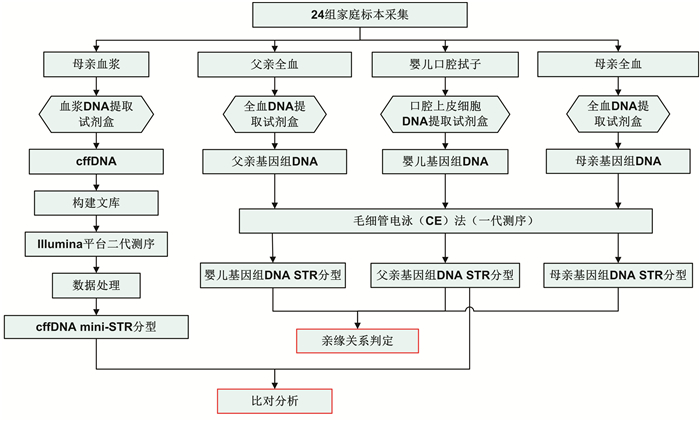
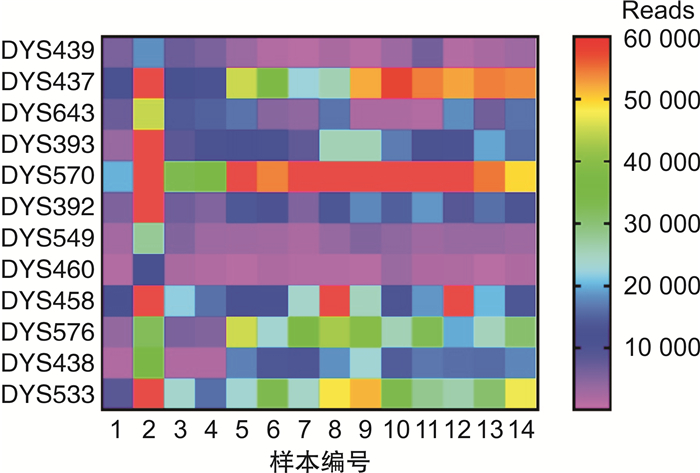
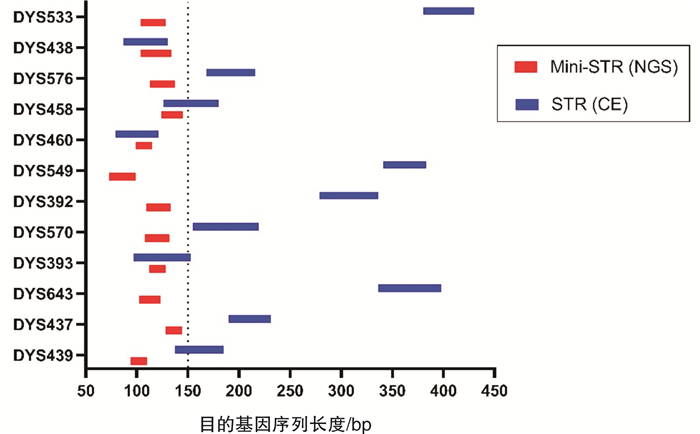
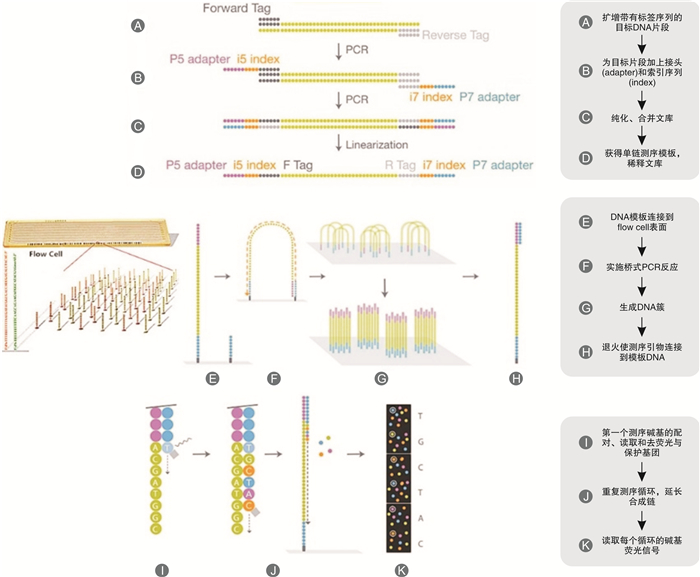
摘要: 目的 评价基于Y-染色体mini-STR的二代测序技术(NGS)通过检测孕妇血浆中胎儿遗传物质完成无创产前亲权鉴定的可行性。方法 从24位孕妇外周血中提取游离胎儿DNA(cffDNA),用Illumina NextSeq 500平台对cffDNA同时进行12个mini-STR基因座的NGS测序分型。cffDNA基因分型结果通过胎儿父亲基因型来验证。对每一组的分型结果计算亲权相关参数。结果 对父亲和出生后的婴儿采用常规的毛细管电泳法(CE)做亲子鉴定,证实了24对父亲和胎儿均具有亲子关系。其中13组男性胎儿的cffDNA所有等位基因均与父亲相同,而1组男性胎儿的cffDNA分型在DYS393出现与父亲不同的等位基因,证实为基因突变;10组女性胎儿的cffDNA均无等位基因检出。利用当地人群单倍型频率可以计算累积亲权指数(CPI)和亲权概率。13组无基因突变的男性胎儿的亲权概率为98.269 9%~99.882 8%,基因突变组的亲权概率为14.871 9%。结论 本研究初步证明了基于NGS的Y-染色体mini-STR用于无创产前亲权鉴定具有较高的准确性,可用于部分案件中在不损伤受害人和胎儿的前提下排除无关男性,并且可以将调查范围扩大到胎儿的所有男性生物学亲属。Abstract: Objectives To evaluate the feasibility of performing non-invasive prenatal paternity testing(NIPPT) by Y-chromosome mini-STR-based next-generation sequencing(NGS).Methods Plasma DNA was extracted from peripheral blood of 24 pregnant women, and cell-free fetal DNA(cffDNA) genotyping at 12 Y-chromosome mini-STR loci was carried out on the Illumina NextSeq 500 system. The cffDNA genotyping result was validated by the paternal genotypes. The paternity testing parameters were calculated for each case.Results The paternity for all 24 family cases was validated by capillary electrophoresis(CE). No missing loci were detected in the 14 male cffDNA mini-STR genotypes by NGS. The alleles of cffDNA and paternal genomic DNA were matched in 13 cases, and a mismatched allele, considered as mutation, appeared at the DYS393 locus in a case. No allele was shown in the 10 female cffDNA. The combined paternity index(CPI) and probability of paternity were calculated according to the local haplotype distribution. The probability of paternity was 98.269 9%-99.882 8% for the cases without mutation, and 14.871 9% for the case with mutation.Conclusion Our study proved that Y-chromosome mini-STR could be used for NGS-based NIPPT with high accuracy, and could be potentially applied for familial searching, paternity exclusion in forensic and sex selection in medical applications.
-

-
表 1 14组男性胎儿cffDNA与其父亲亲权鉴定相关参数
样本编号 单倍型频率 CPI 亲权概率/% 1 0.001 174 852 99.882 8 2 0.017 606 56.8 98.269 9 3 0.001 174 852 99.882 8 4 0.003 521 284 99.649 1 5 0.005 869 170.4 99.416 6 6 0.001 174 852 99.882 8 7 0.003 521 284 99.649 1 8 0.001 174 852 99.882 8 9 0.001 174 852 99.882 8 10 0.003 521 284 99.649 1 11 0.001 174 852 99.882 8 12 0.001 174 852 99.882 8 13 0.002 347 426 99.765 8 141) 0.003 521 0.1746 6 14.869 0 1)存在基因座DYS393等位基因突变。 -
[1] Beta J, Lesmes-Heredia C, Bedetti C, et al. Risk of miscarriage following amniocentesis and chorionic villus sampling: a systematic review of the literature[J]. Minerva Ginecol, 2018, 70(2): 215-219.
[2] Christiansen SL, Jakobsen B, Børsting C, et al. Non-invasive prenatal paternity testing using a standard forensic genetic massively parallel sequencing assay for amplification of human identification SNPs[J]. Int J Legal Med, 2019, 133(5): 1361-1368. doi: 10.1007/s00414-019-02106-0
[3] Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum[J]. Lancet, 1997, 350(9076): 485-487. doi: 10.1016/S0140-6736(97)02174-0
[4] Chang MY, Kim AR, Kim MY, et al. Development of novel noninvasive prenatal testing protocol for whole autosomal recessive disease using picodroplet digital PCR[J]. Sci Rep, 2016, 6: 37153. doi: 10.1038/srep37153
[5] Hyland CA, Millard GM, O'Brien H, et al. Non-invasive fetal RHD genotyping for RhD negative women stratified into RHD gene deletion or variant groups: comparative accuracy using two blood collection tube types[J]. Pathology, 2017, 49(7): 757-764. doi: 10.1016/j.pathol.2017.08.010
[6] Palomaki GE, Best RG. Sequencing Cell-Free DNA in the Maternal Circulation to Screen for Down Syndrome, Other Common Trisomies, and Selected Genetic Disorders[M]. Genomic Applications in Pathology: Springer, 2019: 561-582.
[7] Primacio R, Milot H, Jacob C. Early fetal sex determination using cell-free DNA in micro-volume of maternal plasma[J]. Journal of Pregnancy and Child Health, 2017, 4(6): 1-4.
[8] Ryan A, Baner J, Demko Z, et al. Informatics-based, highly accurate, noninvasive prenatal paternity testing[J]. Genet Med, 2013, 15(6): 473-477. doi: 10.1038/gim.2012.155
[9] Chan KC, Zhang J, Hui AB, et al. Size distributions of maternal and fetal DNA in maternal plasma[J]. Clin Chem, 2004, 50(1): 88-92. doi: 10.1373/clinchem.2003.024893
[10] Breveglieri G, D'Aversa E, Finotti A, et al. Non-invasive Prenatal Testing Using Fetal DNA[J]. Mol Diagn Ther, 2019, 23(2): 291-299. doi: 10.1007/s40291-019-00385-2
[11] Lo YM, Zhang J, Leung TN, et al. Rapid clearance of fetal DNA from maternal plasma[J]. Am J Hum Genet, 1999, 64(1): 218-224. doi: 10.1086/302205
[12] Kayser M. Forensic use of Y-chromosome DNA: a general overview[J]. Hum Genet, 2017, 136(5): 621-635. doi: 10.1007/s00439-017-1776-9
[13] Roewer L. Y-chromosome short tandem repeats in forensics—Sexing, profiling, and matching male DNA[J]. Wiley Interdisciplinary Reviews: Forensic Science, 2019, 1(4): e1336.
[14] Roewer L, Andersen MM, Ballantyne J, et al. DNA commission of the International Society of Forensic Genetics(ISFG): Recommendations on the interpretation of Y-STR results in forensic analysis[J]. Forensic Sci Int Genet, 2020, 48: 102308. doi: 10.1016/j.fsigen.2020.102308
[15] Tam J, Chan YM, Tsang SY, et al. Noninvasive prenatal paternity testing by means of SNP-based targeted sequencing[J]. Prenat Diagn, 2020, 40(4): 497-506.
[16] Barra GB, Santa Rita TH, Chianca CF, et al. Fetal male lineage determination by analysis of Y-chromosome STR haplotype in maternal plasma[J]. Forensic Sci Int Genet, 2015, 15: 105-110. doi: 10.1016/j.fsigen.2014.11.006
[17] Bai X, Li S, Cong B, et al. Construction of two fluorescence-labeled non-combined DNA index system miniSTR multiplex systems to analyze degraded DNA samples in the Chinese Han Population[J]. Electrophoresis, 2010, 31(17): 2944-2948. doi: 10.1002/elps.201000163
[18] Biesecker LG, Bailey-Wilson JE, Ballantyne J, et al. Epidemiology. DNA identifications after the 9/11 World Trade Center attack[J]. Science, 2005, 310(5751): 1122-1123. doi: 10.1126/science.1116608
[19] Rolf B, Keil W, Brinkmann B, et al. Paternity testing using Y-STR haplotypes: assigning a probability for paternity in cases of mutations[J]. Int J Legal Med, 2001, 115(1): 12-15. doi: 10.1007/s004140000201
[20] Peterson RW. A few things you should know about paternity tests(but were afraid to ask)[J]. Santa Clara L Rev, 1982, 22: 667.
[21] Guo F. Population genetics for 17 Y-STR loci in Northern Han Chinese from Liaoning Province, Northeast China[J]. Forensic Sci Int Genet, 2017, 29: e35-e37. doi: 10.1016/j.fsigen.2017.04.012
[22] Thompson R, Zoppis S, McCord B. An overview of DNA typing methods for human identification: past, present, and future[J]. MethodsMol Biol, 2012, 830: 3-16.
[23] Butler JM, Buel E, Crivellente F, et al. Forensic DNA typing by capillary electrophoresis using the ABI Prism 310 and 3100 genetic analyzers for STR analysis[J]. Electrophoresis, 2004, 25(10-11): 1397-412.
[24] Børsting C, Morling N. Next generation sequencing and its applications in forensic genetics[J]. Forensic Sci Int Genet, 2015, 18: 78-89. doi: 10.1016/j.fsigen.2015.02.002
[25] Aly SM, Sabri DM. Next generation sequencing(NGS): a golden tool in forensic toolkit[J]. Arch Med Sadowej Kryminol, 2015, 65(4): 260-271.
[26] Lo YM, Chiu RW. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidies by maternal plasma nucleic acid analysis[J]. Clin Chem, 2008, 54(3): 461-466. doi: 10.1373/clinchem.2007.100016
[27] Zhang S, Han S, Zhang M, et al. Non-invasive prenatal paternity testing using cell-free fetal DNA from maternal plasma: DNA isolation and genetic marker studies[J]. Leg Med(Tokyo), 2018, 32: 98-103. doi: 10.1016/j.legalmed.2018.03.009
[28] Jiang H, Xie Y, Li X, et al. Noninvasive Prenatal Paternity Testing(NIPAT)through Maternal Plasma DNA Sequencing: A Pilot Study[J]. PLoS One, 2016, 11(9): e0159385. doi: 10.1371/journal.pone.0159385
[29] Morling N, Allen RW, Carracedo A, et al. Paternity Testing Commission of the International Society of Forensic Genetics: recommendations on genetic investigations in paternity cases[J]. Forensic Sci Int, 2002, 129(3): 148-157. doi: 10.1016/S0379-0738(02)00289-X
[30] Zhou Z, Shao C, Xie J, et al. Genetic polymorphism and phylogenetic analyses of 21 non-CODIS STR loci in a Chinese Han population from Shanghai[J]. Mol Geneti Genomic Med, 2020, 8(2): e1083. http://onlinelibrary.wiley.com/doi/full/10.1002/mgg3.1083
[31] Gjertson DW, Brenner CH, Baur MP, et al. ISFG: Recommendations on biostatistics in paternity testing[J]. Forensic Sci Int Genet, 2007, 1(3-4): 223-231. doi: 10.1016/j.fsigen.2007.06.006
[32] Morling N, Allen RW, Carracedo A, et al. Paternity Testing Commission of the International Society of Forensic Genetics: recommendations on genetic investigations in paternity cases[J]. Forensic Sci Int, 2002, 129(3): 148-157. doi: 10.1016/S0379-0738(02)00289-X
[33] Ellegren H. Microsatellites: simple sequences with complex evolution[J]. Nat Rev Genet, 2004, 5(6): 435-445. doi: 10.1038/nrg1348
[34] de Jong A, Dondorp WJ, de Die-Smulders CE, et al. Non-invasive prenatal testing: ethical issues explored[J]. Eur J Hum Genet, 2010, 18(3): 272-277. doi: 10.1038/ejhg.2009.203
-




 下载:
下载: