Mutation diagnosis and pathogenic mechanism analysis of the F8 gene in 65 patients with severe haemophilia A in Shanxi province in China
-
摘要: 目的 对65例已确诊的重型血友病A(haemophilia A,HA)患者进行基因变异分析,并探讨其分子发病机制。方法 选取确诊为HA的男性患者65例作为研究对象,FⅧ∶C均小于1%。采用IS-PCR进行内含子22倒位及内含子1倒位筛查,对33例HA倒位阴性患者进行二代测序分析全部外显子序列,确定其变异位点。使用PolyPhen-2、SIFT、Mutation Taster等生物信息学软件预测变异致病性,并分析氨基酸保守性。使用PyMol 2.6软件模拟变异蛋白质的结构。结果 IS-PCR共检测到29例(44.62%)内含子22倒位阳性和3例(4.62%)内含子1倒位阳性。测序结果显示24例患者共检测到27个F8变异位点,包括12例(18.46%)错义突变、8例(12.31%)无义突变、4例(6.15%)缺失突变和3例(4.62%)重复突变,其中3例患者各检测出2个不同的错义突变,其余9例未检测到致病变异,可能为其他特殊突变。共发现c.938T>C、c.3893G>A、c.920T>A、c.2933C>G、c.2669delT、c.4630delC、c.4784delC七种未报道过的新型变异,均为重型HA致病基因。结论 重型HA常见的突变类型仍然是内含子22倒位,本研究新发现的7种HA变异将丰富F8基因变异谱,为今后探讨HA的发病机制提供了基础,明确HA基因诊断为患者提供遗传咨询及临床治疗具有重要意义。Abstract: Objective To analyze the genetic variants and investigate the molecular pathogenesis of 65 patients diagnosed with severe haemophilia A(HA).Methods A total of 65 male patients diagnosed with severe HA and FⅧ∶C levels less than 1%, were selected for this study. IS-PCR was used to screen for intron 22 inversions and intron 1 inversions, and 33 HA inversion-negative patients were analyzed by second-generation sequencing for all exon sequences to identify their variant sites. Bioinformatics software such as PolyPhen-2, SIFT, and Mutation Taster were used to predict variant pathogenicity and analyze amino acid conservation. Furthermore, PyMol 2.6 software was used to simulate the structure of the mutated proteins.Results IS-PCR detected intron 22 inversion in 29 cases(44.62%) and intron 1 inversion in 3 cases(4.62%). The sequencing results revealed a total of 27 F8 mutation sites in 24 patients. This included 12 cases (18.46%) of missense mutations, 8 cases(12.31%) of nonsense mutations, 4 cases(6.15%) of deletion mutations, and 3 cases(4.62%) of duplication mutations. Among these, 3 patients were found to have 2 different missense mutations each. In the remaining 9 cases, no pathogenic variants were detected, suggesting the presence of other unique mutations. Additionally, seven novel and unreported variants were identified, namely c.938T>C, c.3893G>A, c.920T>A, c.2933C>G, c.2669delT, c.4630delC, and c.4784delC. These mutations were all associated with severe HA and were considered pathogenic.Conclusion The most common mutation type of HA remains intron 22 inversion. The identification of seven new HA variants in this study will expand the range of F8 gene mutations, laying the groundwork for further research into the development of HA. This research is crucial for improving the accuracy of HA gene diagnosis, which in turn will enhance genetic counseling and clinical management for affected patients.
-
Key words:
- haemophilia A /
- factor Ⅷ /
- F8 gene /
- genetic variant
-

-
表 1 IS-PCR检测IVS22及IVS1引物序列
引物名称 引物序列(5’-3’) IVS22 ID ACATACGGTTTAGTCACAAGT ED TCCAGTCACTTAGGCTCAG IU CCTTTCAACTCCATCTCCAT IVS1 1-ID GCCGATTGCTTATTTATATC 1-IU TCTGCAACTGGTACTCATC 1-ED GCCTTTACAATCCAACACT 表 2 24例重型HA患者NGS结果
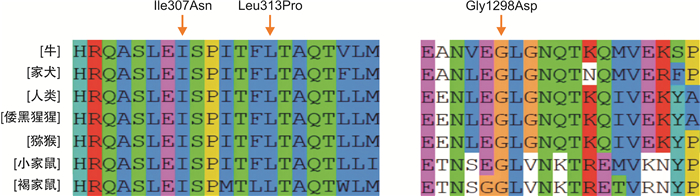
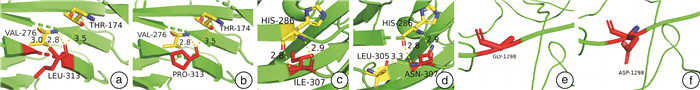
患者编号 年龄/岁 突变类型 外显子编号 突变结构域 核苷酸变化 氨基酸变化 1 39 错义突变 14 B c.3169G>A p.E1057K 错义突变 7 A1 c.938T>C p.L313P 2 31 错义突变 3 A1 c.275G>T p.G92V 3 19 错义突变 18 A3 c.5879G>T p.R1960L 4 7 错义突变 5 A1 c.670G>A p.G224R 5 20 无义突变 14 B c.2933C>G p.S978X 6 22 无义突变 5 A1 c.625C>T p.Q209X 7 1 错义突变 14 B c.3893G>A p.G1298D 错义突变 19 A3 c.6047G>C p.R2016P 8 4 无义突变 16 A3 c.5515C>T p.Q1839X 9 3 无义突变 9 A2 c.1336C>T p.R446X 10 17 无义突变 18 A3 c.5878C>T p.R1960X 11 2 无义突变 14 B c.2404C>T p.Q802X 12 19 错义突变 14 a2 c.2167G>A p.A723T 错义突变 14 B c.3780C>G p.D1260E 13 2 无义突变 22 C1 c.6403C>T p.R2135X 14 16 错义突变 7 A1 c.920T>A p.I307N 15 35 缺失突变 12 B c.4630delC p.L1544Ffs*23 16 19 缺失突变 14 B c.4784delC p.P1595Qfs*26 17 10 无义突变 14 B c.2669delT p.L890X 18 36 缺失突变 12 A2 c.1845delC p.P615fs*45 19 25 重复突变 14 B c.4379dupA p.N1460Kfs*1 20 23 缺失突变 14 B c.3637delA p.I1213Ffs*5 21 18 重复突变 14 B c.3637dupA p.I1213Nfs*28 22 43 重复突变 17 A3 c.5592dupA p.D1865Rfs*16 23 30 错义突变 7 A1 c.907G>C p.A303P 24 2 错义突变 8 a1 c.1063C>T p.R355Y 未曾报道过的F8突变以黑色加粗字体显示。 -
[1] Song X, Zhong J, Xue F, et al. An overview of patients with haemophilia A in China: Epidemiology, disease severity and treatment strategies[J]. Haemophilia, 2020, 27(1): 51-9.
[2] 夏爱丹, 林素仙, 黄瑛, 等. 血友病A患者因子Ⅷ抑制物形成后出血的治疗体会[J]. 中外医学研究, 2013, 11(21): 195-6.
[3] Callaghan MU, Sidonio R, Pipe SW. Novel therapeutics for hemophilia and other bleeding disorders[J]. Blood, 2018, 132(1): 23-30. doi: 10.1182/blood-2017-09-743385
[4] 王稳, 崔东艳, 张艾, 等. 1例女性杂合变异致轻型血友病A的发病机制分析并文献复习[J]. 临床血液学杂志, 2023, 36(7): 528-532. doi: 10.13201/j.issn.1004-2806.2023.07.013
[5] Perrin GQ, Herzog RW, Markusic DM. Update on clinical gene therapy for hemophilia[J]. Blood, 2019, 133(5): 407-14. doi: 10.1182/blood-2018-07-820720
[6] Qu Y, Nie X, Yang Z, et al. The prevalence of hemophilia in mainland China: a systematic review and meta-analysis[J]. Southeast Asian J Trop Med Public Health, 2014, 45(2): 455-66.
[7] Iorio A, Stonebraker J, Chambost H, et al. Establishing the Prevalence and Prevalence at Birth of Hemophilia in Males[J]. Ann Intern Med, 2019, 171(8): 540-546. doi: 10.7326/M19-1208
[8] 张磊, 代新岳. 血友病基因治疗临床研究进展[J]. 临床血液学杂志, 2022, 35(7): 464-468. doi: 10.13201/j.issn.1004-2806.2022.07.003
[9] Al-Allaf FA, Abduljaleel Z, Bogari NM, et al. Identification of six novel factor ⅷ gene variants using next generation sequencing and molecular dynamics simulation[J]. Acta Biochim Pol, 2019, 66(1): 23-31.
[10] Pshenichnikova O, Salomashkina V, Poznyakova J, et al. Spectrum of Causative Mutations in Patients with Hemophilia A in Russia[J]. Genes, 2023, 14(2): 260. doi: 10.3390/genes14020260
[11] 中华医学会血液学分会血栓与止血学组, 中国血友病协作组. 血友病诊断与治疗中国专家共识(2017年版)[J]. 中华血液学杂志, 2017, 38(5): 364-370.
[12] Rossetti LC, Radic CP, Larripa IB, et al. Developing a new generation of tests for genotyping hemophilia-causative rearrangements involving int22h and int1h hotspots in the factor Ⅷ gene[J]. J Thromb Haemost, 2008, 6(5): 830-836.
[13] Beth CM, Haskins ME, Ping W, et al. Successful Phenotype Improvement following Gene Therapy for Severe Hemophilia A in Privately Owned Dogs[J]. PLoS One, 2016, 11(3): e0151800.
[14] Swystun LL, James P. Using genetic diagnostics in hemophilia and von Willebrand disease[J]. Am Soc Hematol, 2015: 152-159.
[15] Marchesini E, Morfini M, Valentino L. Recent Advances in the Treatment of Hemophilia: A Review[J]. Biologics, 2021, 15(6): 221-235.
[16] Manderstedt E, Lind-Halldén C, Ljung R, et al. Droplet digital PCR and mile-post analysis for the detection of F8 int1h inversions[J]. J Thromb Haemost, 2021, 19(3): 732-737.
[17] Gouw SC, Van dB, HM, Oldenburg J, et al. F8 gene mutation type and inhibitor development in patients with severe hemophilia A: systematic review and meta-analysis[J]. Blood, 2012, 119(12): 2922-2934.
[18] Feng Y, Li Q, Shi P, et al. Mutation analysis in the F8 gene in 485 families with haemophilia A and prenatal diagnosis in China[J]. Haemophilia, 2020, 27(1): e88-e92.
[19] Pan TY, Wang CC, Shih CJ, et al. Genotyping of intron 22 inversion of factor Ⅷ gene for diagnosis of hemophilia A by inverse-shifting polymerase chain reaction and capillary electrophoresis[J]. Anal Bioanal Chem, 2014, 406(22): 5447-5454.
[20] El-Maarri O, Singer H, Klein C, et al. Lack of F8 mRNA: a novel mechanism leading to hemophilia A[J]. Blood, 2006, 107(7): 2759-2765.
[21] Johnsen JM, Fletcher SN, Huston H, et al. Novel approach to genetic analysis and results in 3000 hemophilia patients enrolled in the My Life, Our Future initiative[J]. Blood Adv, 2017, 1(13): 824-834.
[22] Chunlei Z, Huihui L, Shuguang Y, et al. Molecular basis of LMAN1 in coordinating LMAN1-MCFD2 cargo receptor formation and ER-to-Golgi transport of FⅤ/FⅧ[J]. Blood, 2010, 116(25): 5698-5706.
[23] Mazurkiewicz-Pisarek A, Płucienniczak G, Ciach T, et al. The factor Ⅷ protein and its function[J]. Acta Biochim Pol, 2016, 26(1): 69-73.
-

计量
- 文章访问数: 248
- 施引文献: 0



 下载:
下载: