-
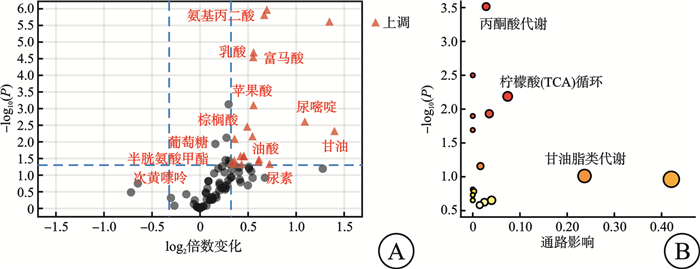
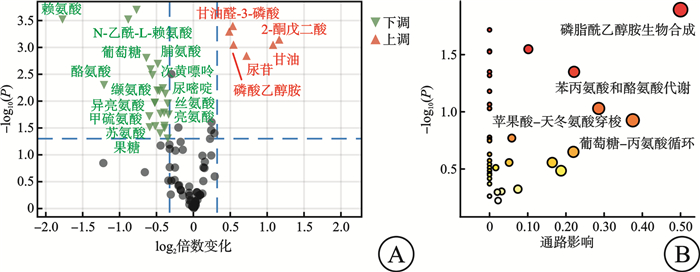
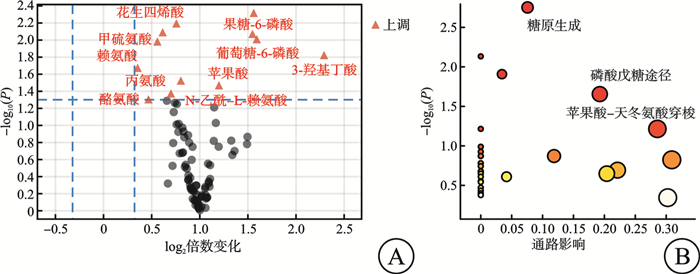
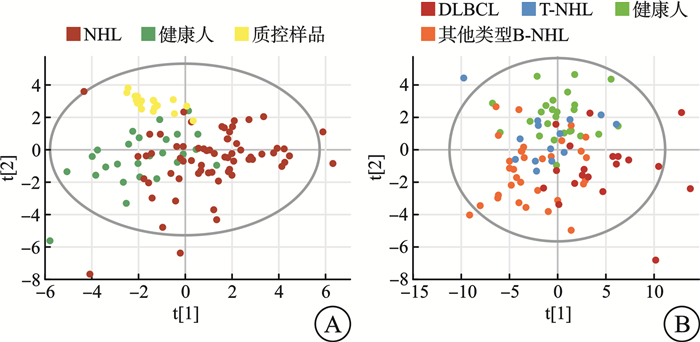
摘要: 目的 探索非霍奇金淋巴瘤(non-Hodgkin lymphoma,NHL)中弥漫大B细胞淋巴瘤(diffuse large B-cell lymphoma,DLBCL)患者与其他类型NHL患者的临床、实验室检查的特征和代谢差异。方法 2018年8月—2023年6月在南京大学医学院附属鼓楼医院收取新鲜组织97例,其中健康人淋巴结增生组织26例,NHL淋巴瘤组织71例。该研究对71例NHL患者的临床资料及实验室特征进行了回顾性分析。同时采用气相色谱-质谱分析(GC/MS)方法,对健康人淋巴结增生组织及NHL淋巴瘤组织进行了非靶向代谢组学分析,并对NHL及DLBCL、其他类型B细胞非霍奇金淋巴瘤(B-cell non-Hodgkin lymphoma,B-NHL)、T细胞非霍奇金淋巴瘤(T-cell non-Hodgkin lymphoma,T-NHL)与健康人之间的具体差异代谢物进行了通路富集分析。结果 71例NHL患者淋巴瘤组织与26例健康人淋巴结增生组织2组间的年龄(P=0.439)及性别(P=0.069)分布差异无统计学意义。71例NHL患者的临床及实验室特征回顾性分析结果显示:DLBCL患者与其他类型B-NHL及T-NHL患者之间在年龄、性别、分期、国际预后指数(International Prognostic Index,IPI)评分等方面差异无统计学意义,但DLBCL较其他类型B-NHL的治疗效果及生存情况不佳。DLBCL与其他类型B-NHL和T-NHL患者比较,除C反应蛋白水平差异有统计学意义外,其余血常规及生化指标亦差异无统计学意义。与其他类型B-NHL比较,DLBCL易出现CD30、MYC、BCL6、MUM1和Ki67的阳性表达,两者之间差异有统计学意义(P<0.05);与T-NHL比较,DLBCL易出现CD79a、MYC、BCL2和MUM1的阳性表达,两者之间差异有统计学意义(P<0.05)。非靶向代谢组学分析发现:NHL患者与健康人存在明显的代谢差异,且DLBCL与其他类型B-NHL和T-NHL之间也存在明显的代谢差异。与健康人比较,DLBCL组织中有12个上调的差异代谢物,主要富集在丙酮酸、柠檬酸循环及甘油脂类代谢途径上;其他类型B-NHL组织中共有19个差异代谢物,其中有14个下调代谢物和5个上调代谢物,富集在磷脂酰乙醇胺生物合成、苯丙氨酸和酪氨酸代谢、苹果酸-天冬氨酸穿梭及葡萄糖-丙氨酸循环代谢途径上;而T-NHL有10个上调的差异代谢物,富集在苹果酸-天冬氨酸穿梭、糖原生成及磷酸戊糖途径代谢途径上。结论 该研究回顾了NHL的临床及实验室特征,并为NHL/DLBCL的代谢性发病机制提供了新的线索,为其临床诊断和药物开发提供了新的思路。Abstract: Objective To explore the features of clinical and laboratory tests, and metabolic differences between patients with diffuse large B-cell lymphoma(DLBCL) in non-Hodgkin lymphoma(NHL) and patients with other types of NHL.Methods From August 2018 to June 2023, 97 fresh tissues were collected from Nanjing Drum Tower Hospital, Affiliated Hospital of Medical School, Nanjing University, including 26 healthy lymph node hyperplasia tissues and 71 NHL lymphoma tissues. The clinical data and laboratory features of 71 patients with NHL were retrospectively analyzed. Meanwhile, the non-targeted metabolomics of hyperplasia lymph node tissues and NHL lymphoma tissues were analyzed by gas chromatography-mass spectrometry(GC/MS), and the pathways enrichment between NHL and DLBCL, other types of NHL and healthy people were analyzed.Results There was no significant difference in age(P=0.439) and sex(P=0.069) between 71 NHL patients and 26 healthy subjects. Retrospective analysis of clinical and laboratory characteristics of 71 NHL patients showed that: there was no statistical difference in age, sex, stage, IPI score between patients with DLBCL and patients with other types of B-cell non-Hodgkin lymphoma(B-NHL) and T-cell non-Hodgkin lymphoma(T-NHL), but DLBCL had poor therapeutic effect and survival compared with other types of B-NHL. Compared with other types of B-NHL and T-NHL patients, DLBCL showed no statistical difference in blood routine and biochemical indexes except for C-reactive protein level. Compared with other types of B-NHL, DLBCL was more likely to show positive expressions of CD30, MYC, BCL6, MUM1 and Ki67, and the difference was statistically significant (P < 0.05). Compared with T-NHL, DLBCL was more likely to show positive expressions of CD79a, MYC, BCL2 and MUM1, and the differences between the two were statistically significant (P < 0.05). Non-targeted metabolomic analysis revealed significant metabolic differences between tissues of NHL patients and healthy individuals, as well as between DLBCL and other types of B-NHL and T-NHL. Compared with healthy people, there were 12 upregulated differential metabolites in DLBCL tissues, which were mainly concentrated in pyruvate, citric acid cycle and glycine metabolism pathway. There were 19 different metabolites in B-NHL tissues, including 14 down-regulated and 5 up-regulated metabolites, which were concentrated in phosphatidylethanolamine biosynthesis, phenylalanine and tyrosine metabolism, malate-aspartate shuttle and glucose-alanine cyclic metabolic pathways. However, T-NHL has 10 upregulated differential metabolites in malate-aspartate shuttle, gluconeogenesis and pentose phosphate pathway.Conclusion This study reviewed the clinical and laboratory features of NHL, and provided new clues for the metabolic pathogenesis of NHL/DLBCL, and provided new ideas for clinical diagnosis and drug development.
-

-
表 1 71例NHL患者的临床特征
例(%) 患者特征 DLBCL
(n=23)其他类型B-NHL
(n=33)T-NHL
(n=15)年龄/岁 64(58~70) 59(52~61) 56(52~65) 性别 女 9(39.1) 17(51.5) 5(33.3) 男 14(60.9) 16(48.5) 10(66.7) 诊断 初诊 19(82.6) 31(93.9) 15(100.0) 复发 4(17.4) 2(6.1) 0 分期 Ⅰ/Ⅱ期 8(34.8) 11(33.3) 3(20.0) Ⅲ/Ⅳ期 15(65.2) 22(66.7) 12(80.0) IPI评分 1~2分 11(47.8) 12(37.5) 8(53.3) 3~4分 12(52.2) 21(62.5) 7(46.7) 化疗 是 18(78.3) 23(69.7) 11(73.3) 否 5(21.7) 10(30.3) 4(26.7) 疗效评价 缓解 9/18(50.0) 16/23(69.6)1) 6/11(54.5) 稳定 0 2/23(8.7)1) 1/11(9.1) 进展 9/18(50.0) 5/23(21.7)1) 4/11(36.4) 生存情况 生存 11(47.8) 24(72.7)1) 5(33.3) 死亡 12(52.2) 9(27.3)1) 10(66.7) 与DLBCL组比较,1) P < 0.05。 表 2 71例NHL患者的实验室检查
X±S 实验室指标 DLBCL(n=23) 其他类型B-NHL(n=33) T-NHL(n=15) 血红蛋白/(g/L) 119.5±23.3 115.0±25.9 120.5±20.0 白细胞/(×109/L) 5.4±1.7 14.2±23.2 6.9±6.0 红细胞/(×1012/L) 4.1±0.7 3.9±0.8 4.2±0.6 血小板计数/(×109/L) 227.5±99.3 185.4±99.9 209.8±120.4 淋巴细胞/(×109/L) 1.2±0.5 9.7±22.4 0.9±0.5 葡萄糖/(mmol/L) 5.2±1.1 5.1±1.8 5.3±1.1 甘油三酯/(mmol/L) 1.2±0.5 1.1±0.8 1.4±0.3 血清总胆固醇/(mmol/L) 3.6±1.2 4.0±1.2 3.5±0.7 白蛋白/(g/L) 38.5±5.1 37.4±4.4 36.1±4.3 谷丙转氨酶/(U/L) 18.9±11.6 18.9±15.5 17.2±6.9 谷草转氨酶/(U/L) 29.6±24.9 23.9±15.9 27.8±14.2 总胆红素/(μmol/L) 10.2±4.9 8.4±4.2 10.5±3.9 直接胆红素/(μmol/L) 2.7±1.1 2.1±0.8 2.9±1.7 乳酸脱氢酶/(U/L) 364.6±298.9 230.2±97.8 359.8±141.6 CRP/(mg/L) 12.8±15.7 11.5±17.0 44.4±57.31) 血肌酐/(μmol/L) 65.8±15.1 61.2±16.1 58.4±14.4 eGFR/(mL/min) 110.1±22.6 112.7±23.0 115.6±31.3 与DLBCL组比较,1)P < 0.05。 -
[1] Liu W, Liu J, Song Y, et al. Burden of lymphoma in China, 2006-2016: an analysis of the Global Burden of Disease Study 2016[J]. J Hematol Oncol, 2019, 12(1): 115. doi: 10.1186/s13045-019-0785-7
[2] Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms[J]. Blood, 2016, 127(20): 2375-2390. doi: 10.1182/blood-2016-01-643569
[3] Ennishi D, Hsi ED, Steidl C, et al. Toward a New Molecular Taxonomy of Diffuse Large B-cell Lymphoma[J]. Cancer Discov, 2020, 10(9): 1267-1281. doi: 10.1158/2159-8290.CD-20-0174
[4] Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas[J]. N Engl J Med, 2008, 359(22): 2313-2323. doi: 10.1056/NEJMoa0802885
[5] Wright GW, Huang DW, Phelan JD, et al. A Probabilistic Classification Tool for Genetic Subtypes of Diffuse Large B Cell Lymphoma with Therapeutic Implications[J]. Cancer Cell, 2020, 37(4): 551-568. e14. doi: 10.1016/j.ccell.2020.03.015
[6] Wang SS. Epidemiology and etiology of diffuse large B-cell lymphoma[J]. Semin Hematol, 2023, 60(5): 255-266. doi: 10.1053/j.seminhematol.2023.11.004
[7] Sehn LH, Salles G. Diffuse Large B-Cell Lymphoma[J]. N Engl J Med, 2021, 384(9): 842-858. doi: 10.1056/NEJMra2027612
[8] Sehn LH, Gascoyne RD. Diffuse large B-cell lymphoma: optimizing outcome in the context of clinical and biologic heterogeneity[J]. Blood, 2015, 125(1): 22-32. doi: 10.1182/blood-2014-05-577189
[9] DeBerardinis RJ, Chandel NS. We need to talk about the Warburg effect[J]. Nat Metab, 2020, 2(2): 127-129. doi: 10.1038/s42255-020-0172-2
[10] Martínez-Reyes I, Cardona LR, Kong H, et al. Mitochondrial ubiquinol oxidation is necessary for tumour growth[J]. Nature, 2020, 585(7824): 288-292. doi: 10.1038/s41586-020-2475-6
[11] Corbet C, Feron O. Cancer cell metabolism and mitochondria: Nutrient plasticity for TCA cycle fueling[J]. Biochim Biophys Acta Rev Cancer, 2017, 1868(1): 7-15. doi: 10.1016/j.bbcan.2017.01.002
[12] Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward[J]. Nat Rev Cancer, 2021, 21(10): 669-680. doi: 10.1038/s41568-021-00378-6
[13] Vasan K, Werner M, Chandel NS. Mitochondrial Metabolism as a Target for Cancer Therapy[J]. Cell Metab, 2020, 32(3): 341-352. doi: 10.1016/j.cmet.2020.06.019
[14] Fessenden M. Metabolomics: Small molecules, single cells[J]. Nature, 2016, 540(7631): 153-155. doi: 10.1038/540153a
[15] Alseekh S, Aharoni A, Brotman Y, et al. Mass spectrometry-based metabolomics: a guide for annotation, quantification and best reporting practices[J]. Nat Meth, 2021, 18(7): 747-756. doi: 10.1038/s41592-021-01197-1
[16] Gooptu M, Whitaker-Menezes D, Sprandio J, et al. Mitochondrial and glycolytic metabolic compartmentalization in diffuse large B-cell lymphoma[J]. Semin Oncol, 2017, 44(3): 204-217. doi: 10.1053/j.seminoncol.2017.10.002
[17] Chen KC, Hsiao IH, Huang YN, et al. Targeting human mitochondrial NAD(P)+-dependent malic enzyme(ME2) impairs energy metabolism and redox state and exhibits antileukemic activity in acute myeloid leukemia[J]. Cell Oncol(Dordr), 2023, 46(5): 1301-1316. doi: 10.1007/s13402-023-00812-x
[18] Pi M, Kuang H, Yue C, et al. Targeting metabolism to overcome cancer drug resistance: A promising therapeutic strategy for diffuse large B cell lymphoma[J]. Drug Resist Updat, 2022, 61: 100822. doi: 10.1016/j.drup.2022.100822
[19] He J, Chen Z, Xue Q, et al. Identification of molecular subtypes and a novel prognostic model of diffuse large B-cell lymphoma based on a metabolism-associated gene signature[J]. J Transl Med, 2022, 20(1): 186. doi: 10.1186/s12967-022-03393-9
[20] Eraslan Z, Papatzikas G, Cazier JB, et al. Targeting Asparagine and Serine Metabolism in Germinal Centre-Derived B Cells Non-Hodgkin Lymphomas(B-NHL)[J]. Cells, 2021, 10(10): 2589. doi: 10.3390/cells10102589
[21] Norberg E, Lako A, Chen PH, et al. Differential contribution of the mitochondrial translation pathway to the survival of diffuse large B-cell lymphoma subsets[J]. Cell Death Differ, 2017, 24(2): 251-262. doi: 10.1038/cdd.2016.116
[22] Caro P, Kishan AU, Norberg E, et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma[J]. Cancer Cell, 2012, 22(4): 547-560. doi: 10.1016/j.ccr.2012.08.014
[23] Zelenetz AD, Gordon LI, Abramson JS, et al. NCCN Guidelines Insights: B-Cell Lymphomas, Version 3.2019[J]. J Natl Compr Canc Netw, 2019, 17(6): 650-661. doi: 10.6004/jnccn.2019.0029
[24] Cheson BD, Fisher RI, Barrington SF, et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification[J]. J Clin Oncol, 2014, 32(27): 3059-3068. doi: 10.1200/JCO.2013.54.8800
[25] Fei F, Zheng M, Xu Z, et al. Plasma Metabolites Forecast Occurrence and Prognosis for Patients With Diffuse Large B-Cell Lymphoma[J]. Front Oncol, 2022, 12: 894891. doi: 10.3389/fonc.2022.894891
[26] Aa JY, Wang GJ, Hao HP, et al. Differential regulations of blood pressure and perturbed metabolism by total ginsenosides and conventional antihypertensive agents in spontaneously hypertensive rats[J]. Acta Pharmacol Sin, 2010, 31(8): 930-937. doi: 10.1038/aps.2010.86
[27] Liu Y, Barta SK. Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment[J]. Am J Hematol, 2019, 94(5): 604-616. doi: 10.1002/ajh.25460
[28] China Anti-cancer Association Lymphoma Committee, Chinese Association for Clinical Oncologists, Medical Oncology Branch of Chinese International Exchange and Promotion Association for Medical and Healthcare. Clinical practice guideline for lympoma in China(2021 Edition)[J]. Chin J Oncol, 2021, 43(7): 707-735.
[29] Liang XJ, Song XY, Wu JL, et al. Advances in Multi-Omics Study of Prognostic Biomarkers of Diffuse Large B-Cell Lymphoma[J]. Int J Biol Sci, 2022, 18(4): 1313-1327. doi: 10.7150/ijbs.67892
[30] Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation[J]. Annu Rev Cell Dev Biol, 2011, 27: 441-464. doi: 10.1146/annurev-cellbio-092910-154237
[31] Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology[J]. Cell, 2017, 168(4): 657-669. doi: 10.1016/j.cell.2016.12.039
[32] Abdel-Wahab AF, Mahmoud W, Al-Harizy RM. Targeting glucose metabolism to suppress cancer progression: prospective of anti-glycolytic cancer therapy[J]. Pharmacol Res, 2019, 150: 104511. doi: 10.1016/j.phrs.2019.104511
[33] Singh AR, Gu JJ, Zhang Q, et al. Metformin sensitizes therapeutic agents and improves outcome in pre-clinical and clinical diffuse large B-cell lymphoma[J]. Cancer Metab, 2020, 8: 10. doi: 10.1186/s40170-020-00213-w
[34] Gonçalves E, Sciacovelli M, Costa ASH, et al. Post-translational regulation of metabolism in fumarate hydratase deficient cancer cells[J]. Metab Eng, 2018, 45: 149-157. doi: 10.1016/j.ymben.2017.11.011
[35] Eniafe J, Jiang S. The functional roles of TCA cycle metabolites in cancer[J]. Oncogene, 2021, 40(19): 3351-3363. doi: 10.1038/s41388-020-01639-8
[36] Schmidt C, Sciacovelli M, Frezza C. Fumarate hydratase in cancer: A multifaceted tumour suppressor[J]. Semin Cell Dev Biol, 2020, 98: 15-25. doi: 10.1016/j.semcdb.2019.05.002
[37] Schmitt A, Xu W, Bucher P, et al. Dimethyl fumarate induces ferroptosis and impairs NF-κB/STAT3 signaling in DLBCL[J]. Blood, 2021, 138(10): 871-884. doi: 10.1182/blood.2020009404
[38] Lim JKM, Leprivier G. The impact of oncogenic RAS on redox balance and implications for cancer development[J]. Cell Death Dis, 2019, 10(12): 955. doi: 10.1038/s41419-019-2192-y
[39] Zhang B, Tornmalm J, Widengren J, et al. Characterization of the Role of the Malate Dehydrogenases to Lung Tumor Cell Survival[J]. J Cancer, 2017, 8(11): 2088-2096. doi: 10.7150/jca.19373
[40] Hanse EA, Ruan C, Kachman M, et al. Cytosolic malate dehydrogenase activity helps support glycolysis in actively proliferating cells and cancer[J]. Oncogene, 2017, 36(27): 3915-3924. doi: 10.1038/onc.2017.36
[41] Guo W, Wang X, Li Y, et al. Function and regulation of lipid signaling in lymphomagenesis: A novel target in cancer research and therapy[J]. Crit Rev Oncol Hematol, 2020, 154: 103071. doi: 10.1016/j.critrevonc.2020.103071
[42] Spiegel RJ, Schaefer EJ, Magrath IT, et al. Plasma lipid alterations in leukemia and lymphoma[J]. Am J Med, 1982, 72(5): 775-782. doi: 10.1016/0002-9343(82)90543-5
[43] Ecker J, Benedetti E, Kindt ASD, et al. The Colorectal Cancer Lipidome: Identification of a Robust Tumor-Specific Lipid Species Signature[J]. Gastroenterology, 2021, 161(3): 910-923. e19. doi: 10.1053/j.gastro.2021.05.009
[44] Vriens K, Christen S, Parik S, et al. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity[J]. Nature, 2019, 566(7744): 403-406. doi: 10.1038/s41586-019-0904-1
[45] Blume-Jensen P, Hunter T. Oncogenic kinase signalling[J]. Nature, 2001, 411(6835): 355-365. doi: 10.1038/35077225
[46] Yang Y, Li S, Wang Y, et al. Protein tyrosine kinase inhibitor resistance in malignant tumors: molecular mechanisms and future perspective[J]. Signal Transduct Target Ther, 2022, 7(1): 329. doi: 10.1038/s41392-022-01168-8
[47] Jiao Q, Bi L, Ren Y, et al. Advances in studies of tyrosine kinase inhibitors and their acquired resistance[J]. Mol Cancer, 2018, 17(1): 36. doi: 10.1186/s12943-018-0801-5
[48] Yang Z, Zheng Y, Gao Q. Lysine lactylation in the regulation of tumor biology[J]. Trends Endocrinol Metab, 2024, 35(8): 720-731. doi: 10.1016/j.tem.2024.01.011
[49] Wiesel-Motiuk N, Assaraf YG. The key roles of the lysine acetyltransferases KAT6A and KAT6B in physiology and pathology[J]. Drug Resist Updat, 2020, 53: 100729. doi: 10.1016/j.drup.2020.100729
[50] Sanderson SM, Gao X, Dai Z, et al. Methionine metabolism in health and cancer: a nexus of diet and precision medicine[J]. Nat Rev Cancer, 2019, 19(11): 625-637. doi: 10.1038/s41568-019-0187-8
-

计量
- 文章访问数: 182
- 施引文献: 0



 下载:
下载: