-
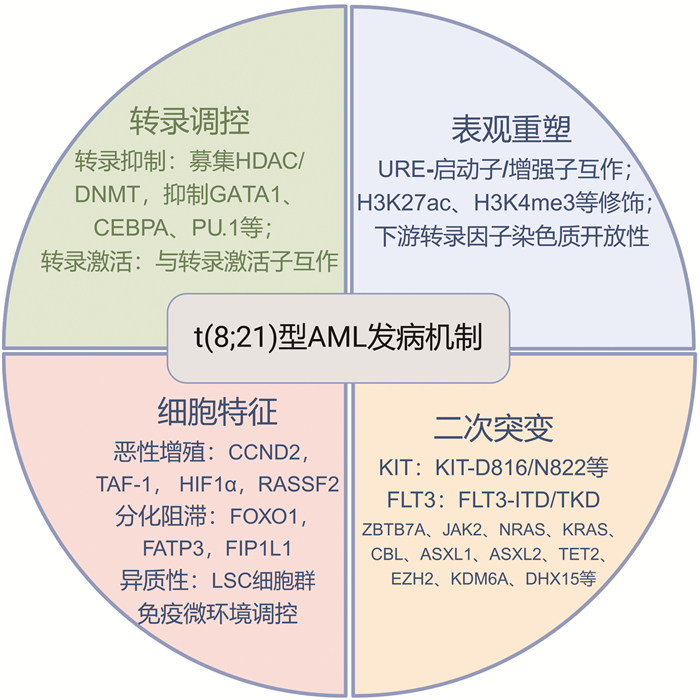
摘要: t(8;21)型急性髓系白血病(AML)是AML中常见的细胞遗传学亚型,以AML1基因和ETO基因重排产生AML1-ETO融合基因为基本特征。近年来,随着该领域基础及临床探索的不断深入,t(8;21)型AML在基因调控、白血病细胞表型特征、协同基因突变、临床异质性等方面的研究均取得新的突破。文章拟就t(8;21)型AML发病机制新进展进行阐述,为临床治疗提供新思路。
-
关键词:
- t(8;21)型急性髓系白血病 /
- 基因调控 /
- 细胞特征 /
- 二次突变
Abstract: t(8; 21) acute myeloid leukemia(AML) is a common cytogenetic subtype of AML, which is characterized by the rearrangement of the AML1 gene and the ETO gene and the formation of the AML1-ETO fusion gene. Recent advances in both basic and clinical research have significantly enhanced our understanding of the mechanisms associated with t(8; 21) AML, including the gene regulation, phenotypic characteristics of leukemia cells, the presence of secondary cytogenetic mutations, and the clinical heterogeneity observed in patients. This article reviews the latest developments in the pathogenesis of t(8; 21) AML, aiming to provide valuable insights into clinical treatment strategies. -

-
[1] 严文伟, 杨崇礼, 杨天楹, 等. 亚急性粒细胞型白血病的临床与诊断[J]. 中华内科杂志, 1964, 12(8): 714-719, 805.
[2] 全国白血病分类分型经验交流讨论会关于白血病分型的建议[J]. 中华血液学杂志, 1980, 1(6): 383-384.
[3] 王志澄. 白血病分类分型讨论会关于急性非淋巴细胞白血病分型的修改建议[J]. 中华血液学杂志, 1987, 8(3): 181.
[4] Rowley JD. Identificaton of a translocation with quinacrine fluorescence in a patient with acute leukemia[J]. Ann Genet, 1973, 16(2): 109-112.
[5] Miyoshi H, Shimizu K, Kozu T, et al. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1 [J]. Proc Natl Acad Sci USA, 1991, 88(23): 10431-10434. doi: 10.1073/pnas.88.23.10431
[6] Miyoshi H, Kozu T, Shimizu K, et al. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript[J]. EMBO J, 1993, 12(7): 2715-2721. doi: 10.1002/j.1460-2075.1993.tb05933.x
[7] 李幼升, 亢非, 周大光, 等. 染色体8;21易位与亚急性粒细胞白血病[J]. 1987, 8(3): 151-152, 155.
[8] 王建祥, 郝玉书, 肖志坚, 等. 急性髓细胞白血病M2b型AML1-ETO融合基因转录本的检测[J]. 中华血液学杂志, 1994, 15(1): 9-11, 52.
[9] 卞寿庚, 严文伟, 李幼升, 等. 亚急性粒细胞白血病是M 2的一个特殊亚型[J]. 中华血液学杂志, 1988, 9(1): 10-14.
[10] 林泽嬉, 卞寿庚, 杨崇礼, 等. M2b白血病细胞的细胞化学染色特征[J]. 中华血液学杂志, 1992, 13(3): 138-140.
[11] Shang L, Chen X, Liu Y, et al. The immunophenotypic characteristics and flow cytometric scoring system of acute myeloid leukemia with t(8;21)(q22;q22);RUNX1-RUNX1T1[J]. Int J Lab Hematol, 2019, 41(1): 23-31. doi: 10.1111/ijlh.12916
[12] Döhner H, Wei AH, Appelbaum FR, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN[J]. Blood, 2022, 140(12): 1345-1377. doi: 10.1182/blood.2022016867
[13] Borthakur G, Ravandi F, Patel K, et al. Retrospective comparison of survival and responses to Fludarabine, Cytarabine, GCSF(FLAG)in combination with gemtuzumab ozogamicin(GO)or Idarubicin(IDA)in patients with newly diagnosed core binding factor(CBF)acute myelogenous leukemia: MD Anderson experience in 174 patients[J]. Am J Hematol, 2022, 97(11): 1427-1434. doi: 10.1002/ajh.26700
[14] Begna KH, Xu X, Gangat N, et al. Core-binding factor acute myeloid leukemia: long-term outcome of 70 patients uniformly treated with"7+3"[J]. Blood Cancer J, 2022, 12(4): 55. doi: 10.1038/s41408-022-00654-0
[15] Yoon JH, Kim HJ, Kim JW, et al. Identification of molecular and cytogenetic risk factors for unfavorable core-binding factor-positive adult AML with post-remission treatment outcome analysis including transplantation[J]. Bone Marrow Transplant, 2014, 49(12): 1466-1474. doi: 10.1038/bmt.2014.180
[16] 中华医学会血液学分会白血病淋巴瘤学组. 成人急性髓系白血病(非急性早幼粒细胞白血病)中国诊疗指南(2023年版)[J]. 中华血液学杂志, 2023, 44(9): 705-712.
[17] Erickson P, Gao J, Chang KS, et al. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1 /ETO with similarity to Drosophila segmentation gene, runt[J]. Blood, 1992, 80(7): 1825-1831. doi: 10.1182/blood.V80.7.1825.1825
[18] Wang J, Hoshino T, Redner RL, et al. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex[J]. Proc Natl Acad Sci USA, 1998, 95(18): 10860-10865. doi: 10.1073/pnas.95.18.10860
[19] Lutterbach B, Westendorf JJ, Linggi B, et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors[J]. Mol Cell Biol, 1998, 18(12): 7176-7184. doi: 10.1128/MCB.18.12.7176
[20] Gelmetti V, Zhang J, Fanelli M, et al. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO[J]. Mol Cell Biol, 1998, 18(12): 7185-7191. doi: 10.1128/MCB.18.12.7185
[21] Liu S, Shen T, Huynh L, et al. Interplay of RUNX1/MTG8 and DNA methyltransferase 1 in acute myeloid leukemia[J]. Cancer Res, 2005, 65(4): 1277-1284. doi: 10.1158/0008-5472.CAN-04-4532
[22] Choi Y, Elagib KE, Delehanty LL, et al. Erythroid inhibition by the leukemic fusion AML1-ETO is associated with impaired acetylation of the major erythroid transcription factor GATA-1[J]. Cancer Res, 2006, 66(6): 2990-2996. doi: 10.1158/0008-5472.CAN-05-2944
[23] Pabst T, Mueller BU, Harakawa N, et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia[J]. Nat Med, 2001, 7(4): 444-451. doi: 10.1038/86515
[24] Buchi F, Masala E, Rossi A, et al. Redistribution of H3K27me3 and acetylated histone H4 upon exposure to azacitidine and decitabine results in de-repression of the AML1/ETO target gene IL3 [J]. Epigenetics, 2014, 9(3): 387-395. doi: 10.4161/epi.27322
[25] Duque-Afonso J, Solari L, Essig A, et al. Regulation of the adaptor molecule LAT2 an in vivo target gene of AML1/ETO(RUNX1/RUNX1T1), during myeloid differentiation[J]. Br J Haematol, 2011, 153(5): 612-622. doi: 10.1111/j.1365-2141.2011.08586.x
[26] Dou L, Yan F, Pang J, et al. Protein lysine 43 methylation by EZH1 promotes AML1-ETO transcriptional repression in leukemia[J]. Nat Commun, 2019, 10(1): 5051. doi: 10.1038/s41467-019-12960-6
[27] Guo M, Chan THM, Zhou Q, et al. Core-binding factor fusion downregulation of ADAR2 RNA editing contributes to AML leukemogenesis[J]. Blood, 2023, 141(25): 3078-3090.
[28] Wang L, Gural A, Sun XJ, et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation[J]. Science, 2011, 333(6043): 765-769. doi: 10.1126/science.1201662
[29] Shia WJ, Okumura AJ, Yan M, et al. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential[J]. Blood, 2012, 119(21): 4953-4962. doi: 10.1182/blood-2011-04-347476
[30] Chen M, Zhu N, Liu X, et al. JMJD1C is required for the survival of acute myeloid leukemia by functioning as a coactivator for key transcription factors[J]. Genes Dev, 2015, 29(20): 2123-2139. doi: 10.1101/gad.267278.115
[31] Ptasinska A, Assi SA, Mannari D, et al. Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding[J]. Leukemia, 2012, 26(8): 1829-1841. doi: 10.1038/leu.2012.49
[32] Ptasinska A, Assi SA, Martinez-Soria N, et al. Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal[J]. Cell Rep, 2014, 8(6): 1974-1988. doi: 10.1016/j.celrep.2014.08.024
[33] Ptasinska A, Pickin A, Assi SA, et al. RUNX1-ETO Depletion in t(8;21) AML Leads to C/EBPα-and AP-1-Mediated Alterations in Enhancer-Promoter Interaction[J]. Cell Rep, 2019, 28(12): 3022-3031.e7. doi: 10.1016/j.celrep.2019.08.040
[34] Nafria M, Keane P, Ng ES, et al. Expression of RUNX1-ETO Rapidly Alters the Chromatin Landscape and Growth of Early Human Myeloid Precursor Cells[J]. Cell Rep, 2020, 31(8): 107691. doi: 10.1016/j.celrep.2020.107691
[35] van der Kouwe E, Heller G, Czibere A, et al. Core-binding factor leukemia hijacks the T-cell-prone PU. 1 antisense promoter[J]. Blood, 2021, 138(15): 1345-1358. doi: 10.1182/blood.2020008971
[36] Trinh BQ, Ummarino S, Zhang Y, et al. Myeloid lncRNA LOUP mediates opposing regulatory effects of RUNX1 and RUNX1-ETO in t(8;21) AML[J]. Blood, 2021, 138(15): 1331-1344. doi: 10.1182/blood.2020007920
[37] Martinez-Soria N, McKenzie L, Draper J, et al. The Oncogenic Transcription Factor RUNX1/ETO Corrupts Cell Cycle Regulation to Drive Leukemic Transformation[J]. Cancer Cell, 2018, 34(4): 626-642.e8. doi: 10.1016/j.ccell.2018.08.015
[38] Xu Y, Man N, Karl D, et al. TAF1 plays a critical role in AML1-ETO driven leukemogenesis[J]. Nat Commun, 2019, 10(1): 4925. doi: 10.1038/s41467-019-12735-z5
[39] Gao XN, Yan F, Lin J, et al. AML1/ETO cooperates with HIF1α to promote leukemogenesis through DNMT3a transactivation[J]. Leukemia, 2015, 29(8): 1730-1740. doi: 10.1038/leu.2015.56
[40] Chen Z, Shao YL, Wang LL, et al. YTHDF2 is a potential target of AML1/ETO-HIF1α loop-mediated cell proliferation in t(8;21) AML[J]. Oncogene, 2021, 40(22): 3786-3798. doi: 10.1038/s41388-021-01818-1
[41] Shao YL, Li YQ, Li MY, et al. HIF1α-mediated transactivation of WTAP promotes AML cell proliferation via m6A-dependent stabilization of KDM4B mRNA[J]. Leukemia, 2023, 37(6): 1254-1267. doi: 10.1038/s41375-023-01904-1
[42] Stoner SA, Liu KTH, Andrews ET, et al. The RUNX1-ETO target gene RASSF2 suppresses t(8;21) AML development and regulates Rac GTPase signaling[J]. Blood Cancer J, 2020, 10(2): 16. doi: 10.1038/s41408-020-0282-9
[43] Breig O, Bras S, Martinez Soria N, et al. Pontin is a critical regulator for AML1-ETO-induced leukemia[J]. Leukemia, 2014, 28(6): 1271-1279. doi: 10.1038/leu.2013.376
[44] Vegi NM, Klappacher J, Oswald F, et al. MEIS2 Is an Oncogenic Partner in AML1-ETO-Positive AML[J]. Cell Rep, 2016, 16(2): 498-507. doi: 10.1016/j.celrep.2016.05.094
[45] Tonks A, Pearn L, Musson M, et al. Transcriptional dysregulation mediated by RUNX1-RUNX1T1 in normal human progenitor cells and in acute myeloid leukaemia[J]. Leukemia, 2007, 21(12): 2495-2505. doi: 10.1038/sj.leu.2404961
[46] Lin S, Ptasinska A, Chen X, et al. A FOXO1-induced oncogenic network defines the AML1-ETO preleukemic program[J]. Blood, 2017, 130(10): 1213-1222. doi: 10.1182/blood-2016-11-750976
[47] Liu X, Liu Y, Rao Q, et al. Targeting Fatty Acid Metabolism Abrogates the Differentiation Blockade in Preleukemic Cells[J]. Cancer Res, 2024, 84(24): 4233-4245. doi: 10.1158/0008-5472.CAN-23-3861
[48] Davis AG, Johnson DT, Zheng D, et al. Alternative polyadenylation dysregulation contributes to the differentiation block of acute myeloid leukemia[J]. Blood, 2022, 139(3): 424-438. doi: 10.1182/blood.2020005693
[49] Jiang L, Li XP, Dai YT, et al. Multidimensional study of the heterogeneity of leukemia cells in t(8;21) acute myelogenous leukemia identifies the subtype with poor outcome[J]. Proc Natl Acad Sci USA, 2020, 117(33): 20117-20126. doi: 10.1073/pnas.2003900117
[50] Liu Y, Liu W, Lai A, et al. Multiomic analysis identifies a high-risk subgroup that predicts poor prognosis in t(8;21) acute myeloid leukemia[J]. Blood Cancer J, 2024, 14(1): 162. doi: 10.1038/s41408-024-01144-1
[51] Schnoeder TM, Schwarzer A, Jayavelu AK, et al. PLCG1 is required for AML1-ETO leukemia stem cell self-renewal[J]. Blood, 2022, 139(7): 1080-1097. doi: 10.1182/blood.2021012778
[52] Kellaway SG, Potluri S, Keane P, et al. Leukemic stem cells activate lineage inappropriate signalling pathways to promote their growth[J]. Nat Commun, 2024, 15(1): 1359. doi: 10.1038/s41467-024-45691-4
[53] Mei Y, Liu Y, Liu W, et al. Identifying ADGRG1 as a specific marker for tumor-reactive T cells in acute myeloid leukemia[J]. Exp Hematol Oncol, 2024, 13(1): 92. doi: 10.1186/s40164-024-00560-0
[54] Li XP, Song JT, Dai YT, et al. Integrative single-cell analysis of longitudinal t(8;21) AML reveals heterogeneous immune cell infiltration and prognostic signatures[J]. Front Immunol, 2024, 15: 1424933.
[55] Fenske TS, Pengue G, Mathews V, et al. Stem cell expression of the AML1/ETO fusion protein induces a myeloproliferative disorder in mice[J]. Proc Natl Acad Sci U S A, 2004, 101(42): 15184-15189. doi: 10.1073/pnas.0400751101
[56] de Guzman CG, Warren AJ, Zhang Z, et al. Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation[J]. Mol Cell Biol, 2002, 22(15): 5506-5517. doi: 10.1128/MCB.22.15.5506-5517.2002
[57] Yuan Y, Zhou L, Miyamoto T, et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations[J]. Proc Natl Acad Sci U S A, 2001, 98(18): 10398-10403. doi: 10.1073/pnas.171321298
[58] Wang YY, Zhao LJ, Wu CF, et al. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice[J]. Proc Natl Acad Sci USA, 2011, 108(6): 2450-2455. doi: 10.1073/pnas.1019625108
[59] Wichmann C, Quagliano-Lo Coco I, Yildiz Ö, et al. Activating c-KIT mutations confer oncogenic cooperativity and rescue RUNX1/ETO-induced DNA damage and apoptosis in human primary CD34+ hematopoietic progenitors[J]. Leukemia, 2015, 29(2): 279-289. doi: 10.1038/leu.2014.179
[60] Christen F, Hoyer K, Yoshida K, et al. Genomic landscape and clonal evolution of acute myeloid leukemia with t(8;21): an international study on 331 patients[J]. Blood, 2019, 133(10): 1140-1151. doi: 10.1182/blood-2018-05-852822
[61] Krauth MT, Eder C, Alpermann T, et al. High number of additional genetic lesions in acute myeloid leukemia with t(8;21)/RUNX1-RUNX1T1: frequency and impact on clinical outcome[J]. Leukemia, 2014, 28(7): 1449-1458. doi: 10.1038/leu.2014.4
[62] Paschka P, Schlenk RF, Weber D, et al. Adding dasatinib to intensive treatment in core-binding factor acute myeloid leukemia-results of the AMLSG 11-08 trial[J]. Leukemia, 2018, 32(7): 1621-1630. doi: 10.1038/s41375-018-0129-6
[63] Dou X, Chen S, Wu D. Prospective Study of Avapritinib in Patients with Relapsed/Refractory or MRD-Positive Core-Binding Factor Acute Myeloid Leukemia with KIT Mutations[J]. Blood, 2024, 144(Supplement 1): 222. doi: 10.1182/blood-2024-207264
[64] Schessl C, Rawat VPS, Cusan M, et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice[J]. J Clin Invest, 2005, 115(8): 2159-2168. doi: 10.1172/JCI24225
[65] Chen X, Venkataraman G, Kline J. AML1/ETO and FLT3-ITD Cooperate to Induce Acute Myeloid Leukemia in Mice[J]. Blood, 2017, 130(Supplement 1): 1399.
[66] Jahn N, Terzer T, Sträng E, et al. Genomic heterogeneity in core-binding factor acute myeloid leukemia and its clinical implication[J]. Blood Adv, 2020, 4(24): 6342-6352. doi: 10.1182/bloodadvances.2020002673
[67] Faber ZJ, Chen X, Gedman AL, et al. The genomic landscape of core-binding factor acute myeloid leukemias[J]. Nat Genet, 2016, 48(12): 1551-1556. doi: 10.1038/ng.3709
[68] Opatz S, Bamopoulos SA, Metzeler KH, et al. The clinical mutatome of core binding factor leukemia[J]. Leukemia, 2020, 34(6): 1553-1562. doi: 10.1038/s41375-019-0697-0
[69] Hartmann L, Dutta S, Opatz S, et al. ZBTB7A mutations in acute myeloid leukaemia with t(8;21) translocation[J]. Nat Commun, 2016, 7: 11733. doi: 10.1038/ncomms11733
[70] Redondo Monte E, Wilding A, Leubolt G, et al. ZBTB7A prevents RUNX1-RUNX1T1-dependent clonal expansion of human hematopoietic stem and progenitor cells[J]. Oncogene, 2020, 39(15): 3195-3205. doi: 10.1038/s41388-020-1209-4
[71] Duployez N, Marceau-Renaut A, Boissel N, et al. Comprehensive mutational profiling of core binding factor acute myeloid leukemia[J]. Blood, 2016, 127(20): 2451-2459. doi: 10.1182/blood-2015-12-688705
[72] Sood R, Hansen NF, Donovan FX, et al. Somatic mutational landscape of AML with inv(16) or t(8;21) identifies patterns of clonal evolution in relapse leukemia[J]. Leukemia, 2016, 30(2): 501-504. doi: 10.1038/leu.2015.141
[73] Thol F, Bollin R, Gehlhaar M, et al. Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications[J]. Blood, 2014, 123(6): 914-920. doi: 10.1182/blood-2013-07-518746
[74] Eisfeld AK, Kohlschmidt J, Schwind S, et al. Mutations in the CCND1 and CCND2 genes are frequent events in adult patients with t(8;21)(q22;q22) acute myeloid leukemia[J]. Leukemia, 2017, 31(6): 1278-1285. doi: 10.1038/leu.2016.332
[75] Meyer T, Jahn N, Lindner S, et al. Functional characterization of BRCC3 mutations in acute myeloid leukemia with t(8;21)(q22;q22.1)[J]. Leukemia, 2020, 34(2): 404-415. doi: 10.1038/s41375-019-0578-6
-

计量
- 文章访问数: 393
- 施引文献: 0



 下载:
下载: